Our team consisted of four mathematics students (Gaspard, Ewan, Alex and Rezwan), and four biochemistry students (Billy, Minulee, Ally and Fiona). The key strengths of the mathematics students were conducting computational analysis on a protein network, analysing protein communities and identifying potential targets for investigation. The primary focus of the biochemistry students was developing hypotheses, investigating the disease/pathway, assessing the biological relevance of proteins found through network theory, and designing appropriate experiments. The differing strengths of each student meant our group needed to collaborate effectively during the semester to ensure that everyone understood the content and was able to contribute to the project productively.
During the early stages of the project, the role of the biochemistry students was to propose a biologically appropriate topic for investigation and the role of the mathematics students was to identify potential proteins of investigation. Initially, we decided to focus on a rare metabolic disorder known as Maple Syrup Urine disease. The biochemistry students identified some obvious proteins involved in the disease pathway and provided the mathematics students with the yeast homologs. This allowed the mathematics students to use community finding algorithms to confirm the proteins did interact and were all present in the same community. Whilst this gave us a reasonable place to start, further investigation of the yeast homologs by the biochemistry students showed that the level of homology was inadequate and we were required to take a different direction.
The biochemistry students determined that the pyruvate metabolism pathway mediated by the pyruvate dehydrogenase complex may be an appropriate pathway to investigate as it was a well-conserved mitochondrial process. The specific area of interest was pyruvate dehydrogenase complex deficiency, a rare metabolic disorder which causes a range of neurological problems. Each week the biochemistry students provided the mathematics students with proteins they had found during their background research into the pathway/disease and the mathematics students ran community finding algorithms to identify any other proteins that may be of particular interest (e.g. proteins that interacted with more than one of the target proteins). Initially, this was completed with the four proteins which were present in the three subunits of the pyruvate dehydrogenase complex. This analysis provided the biochemistry students with a number of kinase and phosphatase proteins that acted to regulate the activity of the PDH complex. Using community finding algorithms, the mathematics students found the seven proteins including the PDH complex proteins and the regulatory proteins separated into two distinct communities. Interacting proteins were also investigated and only two proteins were found to interact with all seven PDH complex and regulatory targets which were PDX1 and PTC6. The mathematics students also determined the statistical importance of each protein in the community by analysing the degree and centrality of the nodes. This information gave the biochemistry students a new perspective on the topic and highlighted the possibility that the kinase and phosphatase proteins could be manipulated to up-regulate the activity of PDH and minimise disease severity.
Through running further mathematical analysis, a close community of proteins was identified which interacted directly with the E2 subunit of the PDH complex. The biochemistry students investigated this community and found the proteins to be involved in metabolism of lipoic acid which ultimately acts as a cofactor for the PDH complex. We decided that this could be an interesting area of investigation with limited previous research. This allowed our group to narrow down the topic of investigation and specifically focus on the cause of pyruvate dehydrogenase complex deficiency and the role of lipoic acid in possible treatment strategies.
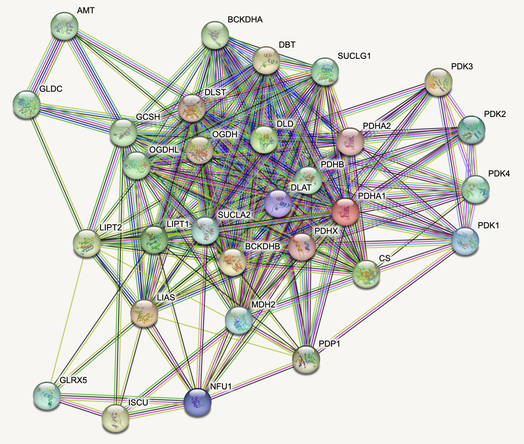
The biochemistry students decided to focus on four proteins involved in the lipoic acid pathway as our primary targets. These proteins were DLAT, LIAS, LIPT1 and NFU1, with yeast homologs LAT1, LIP5, AIM22 and NFU1, respectively. The mathematics students identified other proteins which interacted with either two, three or all of these target proteins. Only two proteins were found to interact with all the target proteins which were LIPT2 and PDP1/2 with yeast homologs LIP2 and PTC5, respectively. This gave the biochemistry students a comprehensive list of proteins which are linked with the lipoic acid pathway and PDH complex and could be investigated within the experimental methods.
During subsequent discussion of the experimental methods, our group decided that we would focus on some biological experiments that could also be modelled through mathematical approaches. For our biochemistry methods, we decided to design one yeast, one human cell line and one mouse experiment. This allowed us to analyse the protein interactions, progression of the disease and possible treatment with lipoic acid in a variety of different models. We also decided to investigate the potential effects of knockouts on the yeast protein communities, so the mathematics students used network methods to visualise this. Further details of these experiments can be found in the experimental methods section.
Overall, whilst the students in our group came from two distinct disciplinary backgrounds, we aimed to apply the methods and expertise of both biochemistry and maths to develop a complex understanding of the protein interactions involved in pyruvate metabolism, pyruvate dehydrogenase complex deficiency and the role of lipoic acid.